-Search query
-Search result
Showing 1 - 50 of 100 items for (author: zhao & dy)

EMDB-18110: 
The fibrillar and amorphous states of polyQ Q97
Method: electron tomography / : Zhao DY

EMDB-18114: 
phagophore in fibrillar polyQ
Method: electron tomography / : Zhao DY
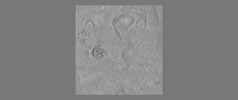
EMDB-18115: 
phagophore and lysosomes with amorphous polyQ
Method: electron tomography / : Zhao DY

EMDB-18116: 
autolysosome, lysosome next to polyQ fibrils are empty
Method: electron tomography / : Zhao DY

EMDB-18117: 
autophagosome and autolysosomes are empty next to fibrillar polyQ
Method: electron tomography / : Zhao DY
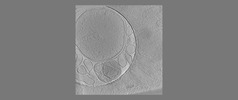
EMDB-18118: 
Isolated autophagosome
Method: electron tomography / : Zhao DY
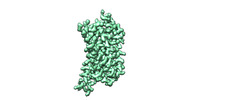
EMDB-43732: 
momSalB bound Kappa Opioid Receptor in complex Gi1
Method: single particle / : Fay JF, Che T

EMDB-43733: 
GR89,696 bound Kappa Opioid Receptor in complex with Gz
Method: single particle / : Fay JF, Che T

EMDB-43734: 
GR89,696 bound Kappa Opioid Receptor in complex with gustducin
Method: single particle / : Fay JF, Che T

EMDB-41876: 
momSalB bound Kappa Opioid Receptor in complex with GoA
Method: single particle / : Fay JF, Che T

EMDB-35410: 
Arabinosyltransferase AftA
Method: single particle / : Gong YC, Rao ZH, Zhang L

EMDB-27804: 
momSalB bound Kappa Opioid Receptor in complex with Gi1
Method: single particle / : Fay JF, Che T

EMDB-27805: 
momSalB bound Kappa Opioid Receptor in complex with GoA
Method: single particle / : Fay JF, Che T

EMDB-27806: 
GR89,696 bound Kappa Opioid Receptor in complex with gustducin
Method: single particle / : Fay JF, Che T

EMDB-27807: 
GR89,696 bound Kappa Opioid Receptor in complex with Gz
Method: single particle / : Fay JF, Che T

PDB-8dzp: 
momSalB bound Kappa Opioid Receptor in complex with Gi1
Method: single particle / : Fay JF, Che T

PDB-8dzq: 
momSalB bound Kappa Opioid Receptor in complex with GoA
Method: single particle / : Fay JF, Che T

PDB-8dzr: 
GR89,696 bound Kappa Opioid Receptor in complex with gustducin
Method: single particle / : Fay JF, Che T

PDB-8dzs: 
GR89,696 bound Kappa Opioid Receptor in complex with Gz
Method: single particle / : Fay JF, Che T

EMDB-33770: 
In situ structure of polymerase complex of mammalian reovirus in the elongation state
Method: single particle / : Bao KY, Zhang XL, Li DY, Zhu P

EMDB-33778: 
In situ structure of polymerase complex of mammalian reovirus in the pre-elongation state
Method: single particle / : Bao KY, Zhang XL, Li DY, Zhu P

EMDB-33779: 
In situ structure of polymerase complex of mammalian reovirus in the reloaded state
Method: single particle / : Bao KY, Zhang XL, Li DY, Zhu P

EMDB-33780: 
In situ structure of polymerase complex of mammalian reovirus in the core
Method: single particle / : Bao KY, Zhang XL, Li DY, Zhu P

EMDB-33787: 
In situ structure of polymerase complex of mammalian reovirus in virion
Method: single particle / : Bao KY, Zhang XL, Li DY, Zhu P

EMDB-33983: 
Structure of CasPi with guide RNA and target DNA
Method: single particle / : Li CP, Wang J, Liu JJ

EMDB-13149: 
Mammalian orthoreovirus T3SA- in complex with the neural receptor NgR1
Method: single particle / : Strebl M, Sutherland DM, Yu X, Hu L, Wang Z, Prasad BVV, Stehle T, Dermody TS

EMDB-13150: 
Mammalian orthoreovirus T3SA-
Method: single particle / : Strebl M, Sutherland DM, Yu X, Hu L, Wang Z, Prasad BVV, Stehle T, Dermody TS

EMDB-32459: 
Composite map of human Kv1.3 channel in apo state with beta subunits
Method: single particle / : Tyagi A, Ahmed T, Jian S, Bajaj S, Ong ST, Goay SSM, Zhao Y, Vorobyov I, Tian C, Chandy KG, Bhushan S

EMDB-32460: 
Composite map of human Kv1.3 channel in dalazatide-bound state with beta subunits
Method: single particle / : Tyagi A, Ahmed T, Jian S, Bajaj S, Ong ST, Goay SSM, Zhao Y, Vorobyov I, Tian C, Chandy KG, Bhushan S

PDB-7wf3: 
Composite map of human Kv1.3 channel in apo state with beta subunits
Method: single particle / : Tyagi A, Ahmed T, Jian S, Bajaj S, Ong ST, Goay SSM, Zhao Y, Vorobyov I, Tian C, Chandy KG, Bhushan S

PDB-7wf4: 
Composite map of human Kv1.3 channel in dalazatide-bound state with beta subunits
Method: single particle / : Tyagi A, Ahmed T, Jian S, Bajaj S, Ong ST, Goay SSM, Zhao Y, Vorobyov I, Tian C, Chandy KG, Bhushan S

EMDB-32486: 
TM domain of human Kv1.3 channel in apo state
Method: single particle / : Tyagi A, Ahmed T, Jian S, Bajaj S, Ong ST, Goay SSM, Zhao Y, Vorobyov I, Tian C, Chandy KG, Bhushan S

EMDB-32487: 
T1 domain of human Kv1.3 channel in apo state with Beta subunit
Method: single particle / : Tyagi A, Ahmed T, Jian S, Bajaj S, Ong ST, Goay SSM, Zhao Y, Vorobyov I, Tian C, Chandy KG, Bhushan S

EMDB-32488: 
TM domain of human Kv1.3 channel in dalazatide-bound state
Method: single particle / : Tyagi A, Ahmed T, Jian S, Bajaj S, Ong ST, Goay SSM, Zhao Y, Vorobyov I, Tian C, Chandy KG, Bhushan S

EMDB-32489: 
T1 domain of human Kv1.3 channel in dalazatide-bound state with Beta subunit
Method: single particle / : Tyagi A, Ahmed T, Jian S, Bajaj S, Ong ST, Goay SSM, Zhao Y, Vorobyov I, Tian C, Chandy KG, Bhushan S

EMDB-32494: 
C1 map of TM domain of human Kv1.3 channel in apo state
Method: single particle / : Tyagi A, Ahmed T, Jian S, Bajaj S, Ong ST, Goay SSM, Zhao Y, Vorobyov I, Tian C, Chandy KG, Bhushan S

EMDB-32495: 
C1 map of TM domain of human Kv1.3 channel in dalazatide-bound state
Method: single particle / : Tyagi A, Ahmed T, Jian S, Bajaj S, Ong ST, Goay SSM, Zhao Y, Vorobyov I, Tian C, Chandy KG, Bhushan S

EMDB-22748: 
SARS-CoV-2 Spike in complex with neutralizing Fab 2B04 (one up, two down conformation)
Method: single particle / : Errico JM, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-22749: 
SARS-CoV-2 Spike RBD in complex with neutralizing Fab 2B04 (local refinement)
Method: single particle / : Errico JM, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-22750: 
SARS-CoV-2 Spike in complex with neutralizing Fab 2H04 (three down conformation)
Method: single particle / : Errico JM, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-22751: 
SARS-CoV-2 Spike RBD in complex with neutralizing Fab 2H04 (local refinement)
Method: single particle / : Errico JM, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-22752: 
SARS-CoV-2 Spike in complex with neutralizing Fab 2B04 (two up, one down conformation)
Method: single particle / : Errico JM, Fremont DH

EMDB-22753: 
SARS-CoV-2 Spike in complex with neutralizing Fab 2H04 (one up, two down conformation)
Method: single particle / : Errico JM, Fremont DH

PDB-7k9h: 
SARS-CoV-2 Spike in complex with neutralizing Fab 2B04 (one up, two down conformation)
Method: single particle / : Errico JM, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-7k9i: 
SARS-CoV-2 Spike RBD in complex with neutralizing Fab 2B04 (local refinement)
Method: single particle / : Errico JM, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-7k9j: 
SARS-CoV-2 Spike in complex with neutralizing Fab 2H04 (three down conformation)
Method: single particle / : Errico JM, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-7k9k: 
SARS-CoV-2 Spike RBD in complex with neutralizing Fab 2H04 (local refinement)
Method: single particle / : Errico JM, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-31148: 
human voltage-gated potassium channel KV1.3
Method: single particle / : Liu S, Zhao Y, Tian C

EMDB-31149: 
human voltage-gated potassium channel KV1.3 H451N mutant
Method: single particle / : Liu S, Zhao Y, Tian C
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
